- language
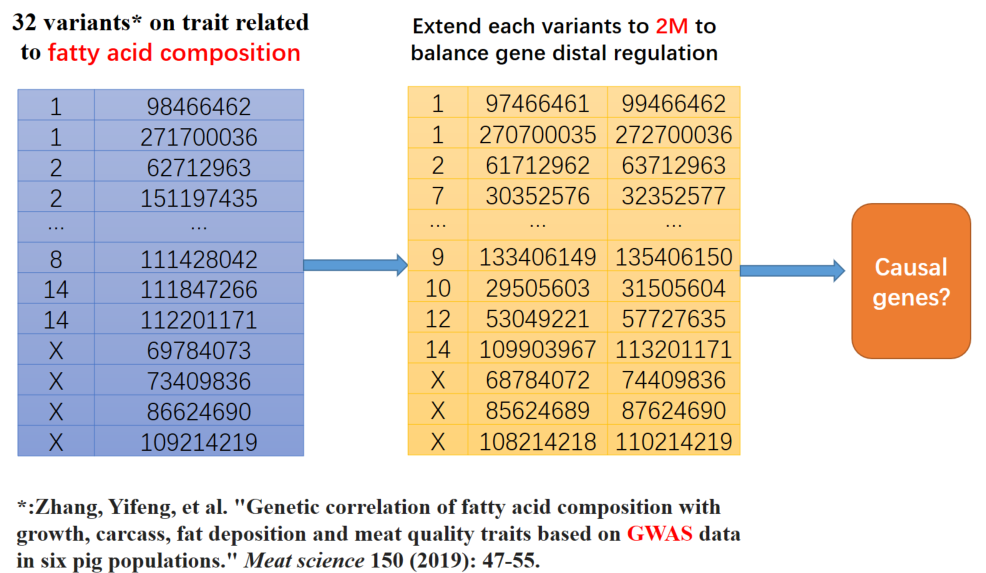
We selected a GWAS literature* containing 32 variants associated with fatty acid composition to demonstrate the use of ISwine.
*:Zhang, Yifeng, et al. "Genetic correlation of fatty acid composition with growth, carcass, fat deposition and meat quality traits based on GWAS data in six pig populations." Meat science 150 (2019): 47-55.
Considering the possible distal regulation, we extended these variants to 2Mb.
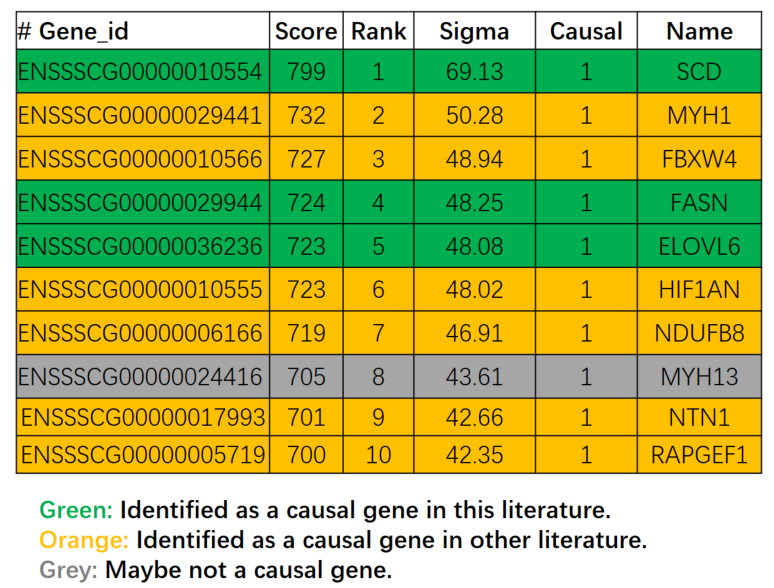
The most effective method of investigation is to consult the literature. We investigated the top 10 genes and found that 9 genes are related to fatty acid composition, including the three causal genes located in this literature.
Sometimes we need more evidence, we can mine some information from the base database (Variation, expression, QTXs).
We selected the top 5 genes to observe the expression level in different tissues and found that the three known causal genes have a very high expression level in fat, and one newly discovered gene (ENSSSCG00000010566) have a high expression level in fat, while another newly discovered gene (ENSSSCG00000029441) have very high expression in muscle, suggesting that the ENSSSCG00000010566 gene may be easier for downstream validation.
We observed the variations of SCD in high fat populations (Asian northern domestic breeds) and low fat populations (European commercial domestic breeds), and found that there are some variations with differences in the promoter region.
ISwine provides all structured data, users can find the genotype of all individuals at any loci, as well as the expression of genes in any sample, so the users can perform the data analysis of interest based on these data.